GraphEx Symposium convenes top network scientists
As the COVID-19 pandemic has shown, we live in a richly connected world, facilitating not only the efficient spread of a virus but also of information and influence. What can we learn by analyzing these connections? This is a core question of network science, a field of research that models interactions across physical, biological, social, and information systems to solve problems.
The Graph Exploitation Symposium (GraphEx), hosted by MIT Lincoln Laboratory, brought together top network science researchers to share the latest advances and applications in the field.
"We explore and identify how exploitation of graph data can offer key technology enablers to solve the most pressing problems our nation faces today," says Edward Kao, a symposium organizer and technical staff in Lincoln Laboratory's AI Software Architectures and Algorithms Group.
The themes of the virtual event revolved around some of the year's most relevant issues, such as analyzing disinformation on social media, modeling the pandemic's spread, and using graph-based machine learning models to speed drug design.
"The special sessions on influence operations and COVID-19 at GraphEx reflect the relevance of network and graph-based analysis for understanding the phenomenology of these complicated and impactful aspects of modern-day life, and also may suggest paths forward as we learn more and more about graph manipulation," says William Streilein, who co-chaired the event with Rajmonda Caceres, both of Lincoln Laboratory.
Social networks
Several presentations at the symposium focused on the role of network science in analyzing influence operations (IO), or organized attempts by state and/or non-state actors to spread disinformation narratives.
Lincoln Laboratory researchers have been developing tools to classify and quantify the influence of social media accounts that are likely IO accounts, such as those willfully spreading false COVID-19 treatments to vulnerable populations.
"A cluster of IO accounts acts as an echo chamber to amplify the narrative. The vulnerable population is then engaging in these narratives," says Erika Mackin, a researcher developing the tool, called RIO or Reconnaissance of Influence Operations.
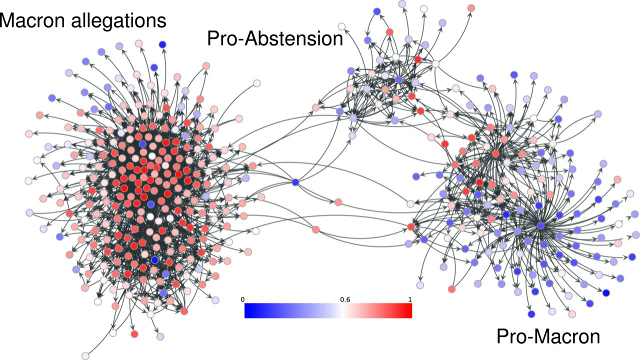
To classify IO accounts, Mackin and her team trained an algorithm to detect probable IO accounts in Twitter networks based on a specific hashtag or narrative. One example they studied was #MacronLeaks, a disinformation campaign targeting Emmanuel Macron during the 2017 French presidential election. The algorithm is trained to label accounts within this network as being IO on the basis of several factors, such as the number of interactions with foreign news accounts, the number of links tweeted, or number of languages used. Their model then uses a statistical approach to score an account's level of influence in spreading the narrative within that network.
The team has found that their classifier outperforms existing detectors of IO accounts, because it can identify both bot accounts and human-operated ones. They've also discovered that IO accounts that pushed the 2017 French election disinformation narrative largely overlap with accounts influentially spreading COVID-19 pandemic disinformation today. "This suggests that these accounts will continue to transition to disinformation narratives," Mackin says.
Pandemic modeling
Throughout the COVID-19 pandemic, leaders have been looking to epidemiological models, which predict how disease will spread, to make sound decisions. Alessandro Vespignani, director of the Network Science Institute at Northeastern University, has been leading COVID-19 modeling efforts in the United States, and shared a keynote on this work at the symposium.
Besides taking into account the biological facts of the disease, such as its incubation period, Vespignani's model is especially powerful in its inclusion of community behavior. To run realistic simulations of disease spread, he develops "synthetic populations" that are built by using publicly available, highly detailed datasets about U.S. households. "We create a population that is not real, but is statistically real, and generate a map of the interactions of those individuals," he says. This information feeds back into the model to predict the spread of the disease.
Today, Vespignani is considering how to integrate genomic analysis of the virus into this kind of population modeling in order to understand how variants are spreading. "It's still a work in progress that is extremely interesting," he says, adding that this approach has been useful in modeling the dispersal of the Delta variant.
As researchers model the virus' spread, Lucas Laird at Lincoln Laboratory is considering how network science can be used to design effective control strategies. He and his team are developing a model for customizing strategies for different geographic regions. The effort was spurred by the differences in COVID-19 spread across U.S. communities, and what the researchers found to be a gap in intervention modeling to address those differences.
As examples, they applied their planning algorithm to three counties in Florida, Massachusetts, and California. Taking into account the characteristics of a specific geographic center, such as the number of susceptible individuals and number of infections there, their planner institutes different strategies in those communities throughout the outbreak duration.
"Our approach irradicates disease in 100 days, but it also is able to do it with much more targeted interventions than any of the global interventions. In other words, you don't have to shut down a full country." Laird adds that their planner offers a "sandbox environment" for exploring intervention strategies in the future.
Machine learning with graphs
Graph-based machine learning is receiving increasing attention for its potential to "learn" the complex relationships between graphical data, and thus extract new insights or predictions about these relationships. This interest has given rise to a new class of algorithms called graph neural networks. Today, graph neural networks are being applied in areas such as drug discovery and material design, with promising results.
"We can now apply deep learning much more broadly, not only to medical images and biological sequences. This creates new opportunities in data-rich biology and medicine," says Marinka Zitnik, an assistant professor at Harvard University who presented her research at GraphEx.
Zitnik's research focuses on the rich networks of interactions between proteins, drugs, disease, and patients, at the scale of billions of interactions. One application of this research is discovering drugs to treat diseases with no or few approved drug treatments, such as for COVID-19. In April, Zitnik's team published a paper on their research that used graph neural networks to rank 6,340 drugs for their expected efficacy against SARS-CoV-2, identifying four that could be repurposed to treat COVID-19.
At Lincoln Laboratory, researchers are similarly applying graph neural networks to the challenge of designing advanced materials, such those that can withstand extreme radiation or capture carbon dioxide. Like the process of designing drugs, the trial-and-error approach to materials design is time-consuming and costly. The Laboratory team is developing graph neural networks that can learn relationships between a material’s crystalline structure and its properties. This network can then be used to predict a variety of properties from any new crystal structure, greatly speeding up the process of screening materials with desired properties for specific applications.
"Graph representation learning has emerged as a rich and thriving research area for incorporating inductive bias and structured priors during the machine learning process, with broad applications such as drug design, accelerated scientific discovery, and personalized recommendation systems," Caceres says.
A vibrant community
Lincoln Laboratory has hosted the GraphEx Symposium annually since 2010, with the exception of last year's cancellation due to COVID-19. "One key take away is that despite the postponement from last year and the need to be virtual, the GraphEx community is as vibrant and active as it's ever been," Streilein says. "Network-based analysis continues to expand its reach and is applied to evermore important areas of science, society, and defense with increasing impact."
In addition to those from Lincoln Laboratory, technical committee members and co-chairs of the GraphEx Symposium included researchers from Harvard University, Arizona State University, Stanford University, Smith College, Duke University, the U.S. Department of Defense, and Sandia National Laboratories.